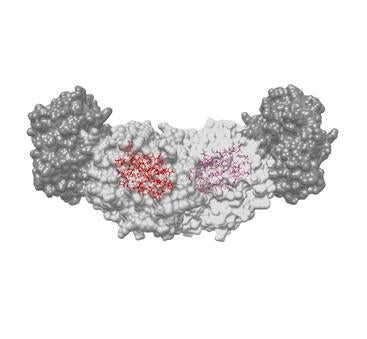
UC Riverside scientists have developed a technique for solving a decades-old mystery involving the chemical in turkey that makes people sleepy. Their ability to map atoms involved in producing tryptophan opens the door to new antibiotic and antifungal drugs.
In addition to its famous post-Thanksgiving side effects, tryptophan performs key functions that many people aren’t aware of. Chiefly, it is a building block for all proteins. Without it, people would have a hard time sleeping, growing or converting food into energy. It is essential not just for humans, but for other organisms like bacteria and fungi as well.
Understanding how to stop disease-causing organisms from making their own tryptophan could enable an entirely new class of treatment drugs.
“The cells in our body don’t make tryptophan – we have to consume it. But bacteria make their own and if that process gets shut down, they will die,” said Jacob Holmes, UCR chemistry graduate student and first author on a paper describing the research.
“So, if we could ingest something that shuts down the enzymes in their bodies making tryptophan, it would not affect our cells but would potentially kill the invasive bacterial cells,” he said.
For more than 20 years, scientists have known about a chemical that stops tryptophan-producing enzymes in cells, called benzimidazole. But until now, they haven’t been able to see how it works. Leonard Mueller, lead principal investigator and chair of UCR’s Chemistry Department, and his colleagues have pioneered new techniques that allow them to do so.
Their work is detailed in a paper published in the Proceedings of the National Academy of Sciences.
Part of the problem with earlier techniques is the inability to see the position of hydrogen atoms. Hydrogen makes up half of the atoms in a protein. Without seeing where they are, creating a true picture of the chemical interactions, and how the molecules fit together, was impossible.
“Imagine you’re browsing a new dating app to match therapeutics with protein targets, and you can only see pixelated avatars of the molecules and their targets. You don’t have enough information to swipe right or left,” Mueller said.
“If you’re trying to design drugs, understanding how other atoms are arranged is useful but you really need to also see the hydrogen atoms to know if there is a match,” he added.
First, the team used a tool involving x-rays to find all the non-hydrogen atoms involved. Next, they made use of atoms’ nuclear magnetism to map out the molecules’ chemical structure, including the hydrogen atom locations. Finally, they used computer modeling to superimpose the images and bring the two techniques together with a resolution that neither could have achieved alone.
“None of these techniques alone can give you the result, but combined, you’re really looking at the bigger picture of chemical reactivity,” said Rittik Ghosh, UCR graduate student in biochemistry and study co-author.
“For so long we’ve been guessing what the active sites in this reaction have looked like. This is one of the first techniques that can bring the chemistry to life,” said Mueller. “We think it will be powerful for designing therapeutics as well as industrial chemical transformations.”